Sunday, October 25, 2009
Others - Aptamers
Saturday, October 24, 2009
Histopathology – Special Stains
HELLOS!
it's yet, my turn again for posting. sorry for the really late posting. hahas.
anyway. i'm currently at the staining department of histopathology lab.. so yup, i'm just gonna share about a few of the manual special stains, as well as about this machine, Ventana NexES special stain, which our lab uses for certain special stain orders. =)
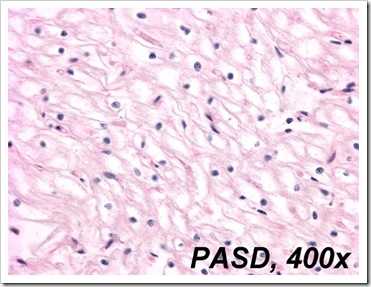
Manual Special stain 1: PAS/D
Also known as, Periodic Acid Schiff Diastase.
It is a stain commonly used to differentiate glycogen from the other carbohydrates.
Procedure:
- Dewax and bring sections to water (i.e, place slides under running water)
- Defroze diastase (which is normally kept in the fridge for storage), after which cover sections with diastase completely for 30 minutes.
- Wash well in water.
- Drip excess water off section, and treat section with 1% periodic acid for 5 minutes.
- Wash well in water.
- Drip excess water off section, and treat section with Schiff's reagent for 3 minutes.
- Wash well in running water.
- Counter stain nuclei with haematoxylin for 1 minute
- Blue in water for 5 minutes, and examine under microscope to check quality of stain.
- Dehydrate and mount slide with Depex.
Any presence of glycogen will be stain magenta on the PAS stained slide while for the PAS/D stained slide, there will be absence of it as glycogen has already been digested by diastase, hence no reaction will occur with schiff's reaction. A control should always be used to display positive results (i.e. sections appear pale pink)
If sections appear as magenta while control section displayed positive result, it shows that there are still presence of glycogen on the section, suggesting possiblilty of glycogen storage disease.
Positive PASD stain. Section stained pale pink instead of magenta.
Negative PASD stain, whereby glycogen are still present on section and stained magenta as seen.
Picture taken from http://path.upmc.edu/cases/case435/images/fig04.jpg and http://emedicine.medscape.com/article/941632-diagnosis respectively.
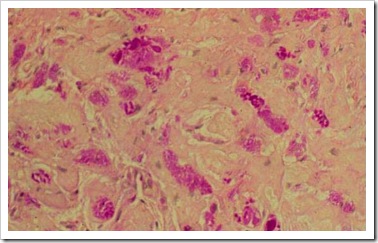
Manual Special Stain 2: Mucicarmine Stain – Southgate’s mucin
Mucicarmine Stain (MC stain), is a type of stain which stains mucin. Mucin is a secretion that is produced by various epithelial cells and connective tissue. When mucin is produced in excess, it may suggest possibility of inflammation of epithelial cells or certain intestinal carcinoma.
Carmine will act as a cationic dye in the presence of aluminium, which will bind with mucin.
Procedure:
- Dewax and bring sections to water (i.e. place slides under running water).
- Treat slides with Weigert Iron Haematoxylin for 10 minutes.
- Wash and blue in water for 5 minutes.
- Stain with mucicarmine solution for 20 minutes.
- Wash well with water
- Dehydrate, and mount slides with depex.
Mucin will be stained red, while nuclei will be stained blue.
Picture taken from http://www.ventanamed.com/includes/galleryThumbnail.php?id=12
Manual Special Stain 3: Victoria Blue Nuclear fast red stain (VB stain)
This is a stain which stains for HBsAg and elastic fibres.
Procedure:
- Dewax and bring sections to water (i.e. place slides under running water).
- Prepare acidified Potassium Permanganate solution
- 6.5 ml of distilled water + 3 ml of 0.5% potassium permanganate + 0.5 ml of 3% sulphuric acid
- Treat sections with acidified potassium permangante solution for 5 minutes.
- Wash well in water.
- Treat sections with 4% sodium metabisulphite for 1 minute.
- Wash well in water.
- Dry slides on hotplate before placing them into Victoria Blue solution for minimum 4 hours, but preferably 24hrs.
- Differentiate in 70% alcohol until background is clear. (Approximately 1minute)
- wash well in water
- Treat section with nuclear fast red stain for 3 minutes.
- Wash well in water
- Dehydrate, and mount slides with depex.
HBsAg, elastic fibres, lipofuchsin and mast cells will be stained blue, while cytoplasm and nuclei will be stained red.
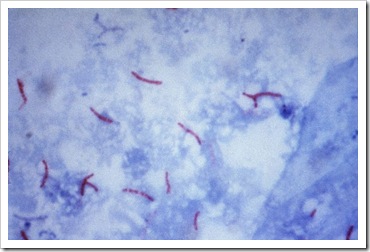
Manual stain 4: Ziehl Neelsen Stain (ZN or TB stain)
This stain is used to demonstrate acid fast bacteria strains belonging to the genus mycobacterium. Lipid capsule of the acid fast organism will taje up carbol-fuchsin and resist decolourization with a dilute acid rinse. This lipid capsule of the mycobacterium will not be stained by methylene blue during bluing. This stain is also commonly known for detection of mycobacterium tuberculosis, which caused tuberculosis in patients, hence is other wise known as Tuberculosis Stain (TB stain)
Procedure:
- Dewax and bring sections to water (i.e. place slides under running water).
- Treat section with commercial TB solution (carbol-fuchsin) for 5 minutes.
- Wash well in water
- Differentiate with 1% acid alcohol until carbol fuchsin is remove from section (i.e. section is clear from stain)
- wash well in water.
- Counterstain with 1% Loeffler’s methylene blue for 10 seconds
- wash well in water.
- Differentiate in 95% alcohol until section turns sky blue colour.
- Dehydrate, and mount slides with depex.
Acid-fast bacteria strains will be stained bright red, while the background will be stained blue.
Picture taken from http://en.wikipedia.org/wiki/File:Mycobacterium_tuberculosis_Ziehl-Neelsen_stain_02.jpg
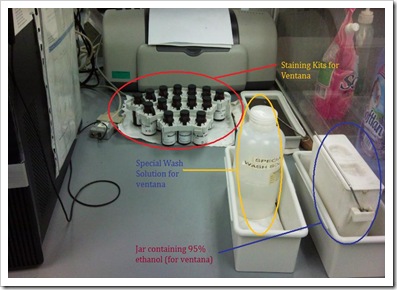
Ventana NexES special stain machine
It is a computerized barcode-driven special stain stainer that automatically applies staining reagents to the microscopic slides and mixed over the entire section. Staining process is pre-programmed in the software and accomplished by the serial application of reagents, all of which are held on the reagent carousel (otherwise known as reagent loading tray).
Liquid coverslip is applied before each application of the reagent to inhibit evaporation. Quantity and amount of reagent to be applied on each slide is also pre-programmed by the software.
Staining reagent kits are supplied in vials with a barcode labelled carrier, which can be fixed onto the reagent carousel of the machine. The computer ‘recognises’ the barcode and carries out the staining procedure accordingly.
Barcode labels are printed from the software, carrying the biopsy number of the specimen/slide, as well as the barcode specific for performing a particular stain.
Before slides are placed into the machine, it must first be labelled with the barcode labels, and dewaxed. After which, slides are placed into the machine and a special wash solution is added to the slides prior to staining. This aids to prevent sections from drying up.
Appropriate staining kits must be loaded to the reagent carousel, with caps uncapped. Before initialising a run, check that waste bottle is still sufficient for usage, and there is sufficient levels of buffered wash solution and liquid coverslip for the run.
When the machine runs the staining program, it will read off the slide and reagent barcode labels, and information for staining will be downloaded from the system to the staining module to proceed run.
When the run has completed, slides are removed from the machine, and the underside of the slides are gently clean with a gauze to remove residue stain solutions. It is then placed into 95% ethanol to remove excess liquid coverslip, afterwhich, proceeded on to dehydration as per normal. Slides will then be mount with depex, and the machine will be cleaned using a special cleaning kit purchased from the company.
Pictures taken with permission from lab.
that’s all for now~
Cheers,
Ang Yu Hui
0702632A