This is gonna be my last entry!
Alright, I was in the bone marrow lab for my last section.
but however, i will not mention about the bone marrow aspiration
as vanessa had done so in her blog post (group 6).
pssst! its my most enjoyable lab experience ever! :D
In the bone marrow lab, not only do we go down to the ward
for bone marrow aspiration, we also do staining of the slides and viewing and
reporting of the results. So in this entry, i'll describe to you guys the different
stainings!
Maygrunwald-Giemsa stain:
it is a romanowsky stain used in morphologic classification
of hematopoietic cells. Giemsa alone can demonstrate malarial parasites in thick film.
Romanowsky stain consists of methylene blue and oxidation product and eosin Y or eosin B.
The combined action of these dyes will produce the Romanowsky effect yielding a purple colour
to nuclei of leukocytes and neutrophil granules, and pink colour to erythrocytes. M/G stained wedged bone marrow smear tells us the cellularity, quantity, quality of the magekaryocytes. Presence of tumour clumps should be noted when viewing slides. It also stains the erythroid and granulocytic cells, lymphoid, reticulum and plasma cells.
Procedure :
1) Add 1:1 ratio of Maygrunwald stain to buffered water onto the slides.
2) Mix well and leave it for 7 minutes.
3) Wash throughly.
4) Add 1ml of Giema stain adn 9ml of buffered water.
5) Mix well and leave it for 12 minutes.
6) Wash and air dry.

picture taken from http://www.bccancer.bc.ca/NR/rdonlyres/5A3C3A86-4549-4422-8258-BE8AE2635461/3748/adenogiantcell03208501w2.jpg
Iron stain :
The HCl caused ferric iron in hemosiderin to release from protein. The free iron react with potassium ferrocyanide and gives an insoluble blue compound called ferric ferrocyanide. The ferritin which is soluble does not give a positive reaction. An increase in iron stores shows more hemosiderin granules that are often in clumps on the slides. Hemosiderin is stained blue-green in colour while nuclei is stained red.
Iron stain procedure :
1) A control is used together with the first iron stain slide of the day.
2) Fix the slides with methanol for 10-20 minutes.
3) Wash it with water
4) Mix 1ml of HCL and 1ml of potassium ferrocyanide into a testtube and mix.
5) Pour the mixture onto the slides and leave it for 15 minutes.
6) Counterstain with neutral red solution for one minute.
7) Wash and air-dry

Picture taken from http://aphlabs.com/yahoo_site_admin/assets/images/DSCN8716.187192835_std.JPG
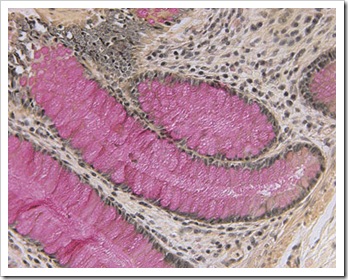
Periodic Acid Schiff stain :
It stains a variety of carbohydrates including glycogen which is often found in haemopoietic cells. It is used to differentiate the diagnosis of acute leukemia. Lymphoblast in coarse granules or large block on clear background are PAS positive. Myeloblast and monoblast are PAS negative. The product will be red in colour. Neutrophils are magenta and cytoplasm are red in colour. Normal erythroblasts do not stain PAS. However PAS is strongly positive in erythroleukemia.
Procedure :
1) Fix the slides with methanol for 15-20 minutes.
2) Wash and add periodic acid and stain it for 10minutes.
3) Rinse and AIR-DRY!
4) Add schiff reagent and leave it for 20 minutes
5) Rinse with water
6) Counterstain with Harris Haematoxylin for 10minutes.
7) Run in tap water for 5minutes.

Picture taken from http://www.kidneypathology.com/Im%E1genes/GNM/05-7007.PAS.2.w.jpg
Peroxidase Stain:
It establish and confirm the diagnosis of acute myeloid leukaemia since lymphoblasts are negative. It requires >3% of bone marrow blasts to show peroxidase activity for acute leukemia to classify as myeloid. Myeloloperoxidase deficiency produces clinical picture similar to that of chronic granulocytic disease. Active myeloperoxidase in granulocytes implies that opsonin function is intact. Cytoplasm shows golden yellow granules with nuclei purple in colour. Monocytes, mature and immature granulocytes and many myeloblasts are positive for peroxidase stain. Lymphocytes, megakaryocytes, plasma cells and erythroblasts are negative.
Procedure :
1)Fix with formol ethanol for 1 min
2) Stain with Ortho-tolidine reagent for 7min
3) rinse under tap water
4) Counterstain wil diluted giemsa for 30mins or more.
Sudan Black stain :
It is clinically used as a marker in differentiation of acute myeloid leukaemia from acute lymphoblastic leukaemia. It is more sensitive more less specific than myeloperoxidase stain in identifying myeloblast. Neutrophils, its precursors and eosinophils are stained blue black. Monocytes stain less intensely than neutrophils. Lymphocytes do not stain with Sudan black.
Positive staining is dark brown to black in cytoplasm of mature and immature granulocytes. Megakaryocytes, platelets, erythroblasts, plasma cells, lymphoblasts are negative.
Neutrophils and precursors are strongly positive. Eosinophils show positivity with appearance. Basophils are variable. Myeloblasts activity usually parallels peroxidase activity with a few positibe granules. Monocytes tend to show fine granular staining.
Procedure:
1) Fix with formol ethanol for one min
2) Stain with sudan black solution for 30mins
3) Rinse in 70% ethanol followed by tap water.
4) counterstain with diluted giemsa for 30mins or more.

Feel free to ask any questions! :D